Week 4: Grouping and summarizing
| dplyr |
|---|
| 1.0.3 |
Summarizing data by group
Let’s first create a dataframe listing the average delay time in minutes, by day of the week and by quarter, for Logan airport’s 2014 outbound flights.
df <- data.frame(
Weekday = factor(rep(c("Mon", "Tues", "Wed", "Thurs", "Fri"), each = 4),
levels = c("Mon", "Tues", "Wed", "Thurs", "Fri")),
Quarter = paste0("Q", rep(1:4, each = 5)),
Delay = c(9.9, 5.4, 8.8, 6.9, 4.9, 9.7, 7.9, 5, 8.8, 11.1, 10.2, 9.3, 12.2,
10.2, 9.2, 9.7, 12.2, 8.1, 7.9, 5.6))The goal will be to summarize the table by Weekday as shown in the following graphic.
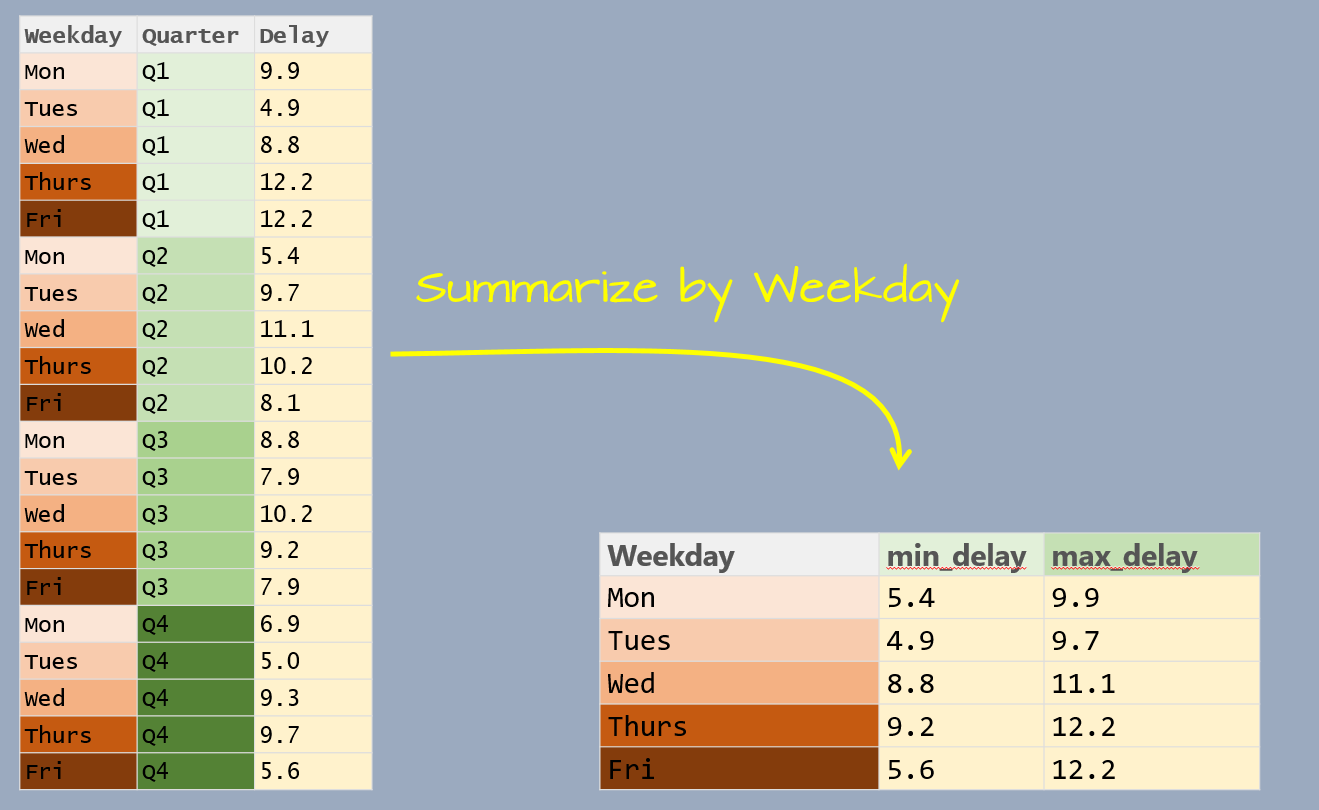
The data table has three variables: Weekday, Quarter and Delay. Delay is the value we will summarize which leaves us with one variable to collapse: Quarter. In doing so, we will compute the Delay statistics for all quarters associated with a unique Weekday value.
This workflow requires two operations: a grouping operation using the group_by function and a summary operation using the summarise function. Here, we’ll compute two summary statistics: minimum delay time and maximum delay time.
library(dplyr)
df %>%
group_by(Weekday) %>%
summarize(min_delay = min(Delay), max_delay = max(Delay))# A tibble: 5 x 3
Weekday min_delay max_delay
* <fct> <dbl> <dbl>
1 Mon 5.4 9.9
2 Tues 4.9 9.7
3 Wed 8.8 11.1
4 Thurs 9.2 12.2
5 Fri 5.6 12.2Note that the weekday follows the chronological order as defined in the Weekday factor.
You’ll also note that the output is a tibble. This data class is discussed at the end of this page.
Grouping by multiple variables
You can group by more than one variable. For example, let’s build another dataframe listing the average delay time in minutes, by quarter, by weekend/weekday and by inbound/outbound status for Logan airport’s 2014 outbound flights.
df2 <- data.frame(
Quarter = paste0("Q", rep(1:4, each = 4)),
Week = rep(c("Weekday", "Weekend"), each=2, times=4),
Direction = rep(c("Inbound", "Outbound"), times=8),
Delay = c(10.8, 9.7, 15.5, 10.4, 11.8, 8.9, 5.5,
3.3, 10.6, 8.8, 6.6, 5.2, 9.1, 7.3, 5.3, 4.4))The goal will be to summarize the delay time by Quarter and by Week type as shown in the following graphic.
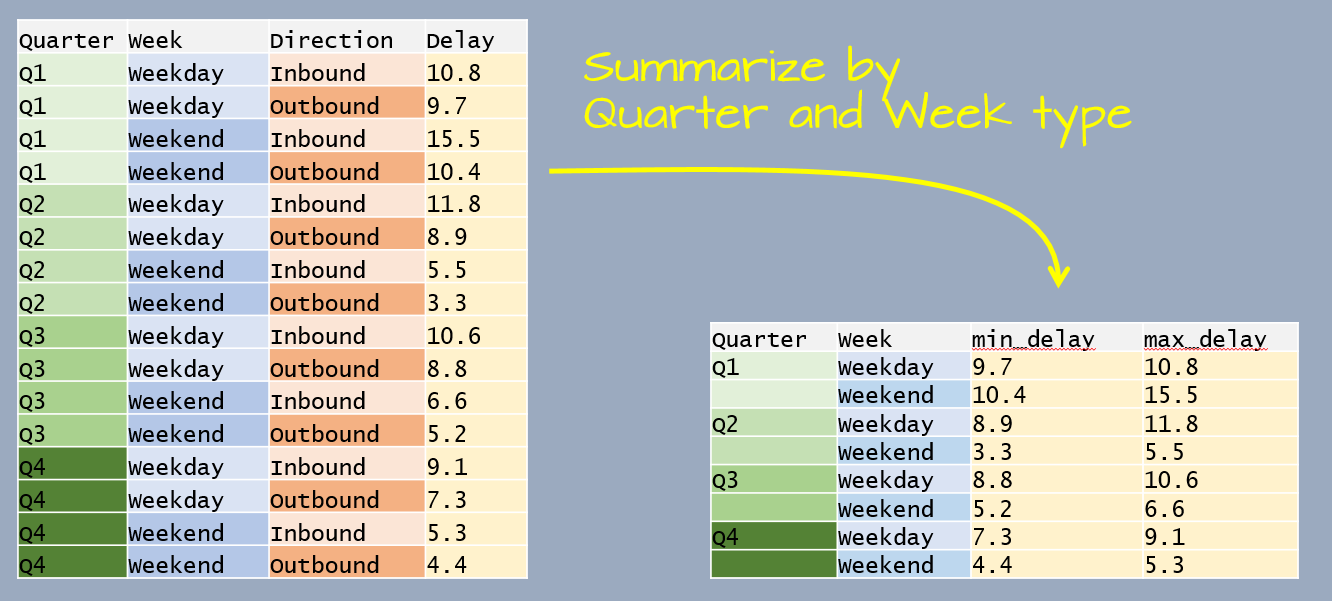
This time, the data table has four variables. We are wanting to summarize by Quater and Week which leaves one variable, Direction, that needs to be collapsed.
df2 %>%
group_by(Quarter, Week) %>%
summarize(min_delay = min(Delay), max_delay = max(Delay))# A tibble: 8 x 4
# Groups: Quarter [4]
Quarter Week min_delay max_delay
<chr> <chr> <dbl> <dbl>
1 Q1 Weekday 9.7 10.8
2 Q1 Weekend 10.4 15.5
3 Q2 Weekday 8.9 11.8
4 Q2 Weekend 3.3 5.5
5 Q3 Weekday 8.8 10.6
6 Q3 Weekend 5.2 6.6
7 Q4 Weekday 7.3 9.1
8 Q4 Weekend 4.4 5.3The following section demonstrates other grouping/summarizing operations on a larger dataset.
A working example
The data file FAO_grains_NA.csv will be used in this exercise. This dataset consists of grain yield and harvest year by North American country. The dataset was downloaded from http://faostat3.fao.org/ in June of 2014.
Run the following line to load the FAO data file into your current R session.
dat <- read.csv("http://mgimond.github.io/ES218/Data/FAO_grains_NA.csv", header=TRUE)Make sure to load the dplyr package before proceeding with the following examples.
library(dplyr)Summarizing by crop type
The group_by function will split any operations applied to the dataframe into groups defined by one or more columns. For example, if we wanted to get the minimum and maximum years from the Year column for which crop data are available by crop type, we would type the following:
dat %>%
group_by(Crop) %>%
summarise(yr_min = min(Year), yr_max=max(Year))# A tibble: 11 x 3
Crop yr_min yr_max
* <chr> <int> <int>
1 Barley 1961 2012
2 Buckwheat 1961 2012
3 Canary seed 1980 2012
4 Grain, mixed 1961 2012
5 Maize 1961 2012
6 Millet 1961 2012
7 Oats 1961 2012
8 Popcorn 1961 1982
9 Rye 1961 2012
10 Sorghum 1961 2012
11 Triticale 1989 2012Count the number of records in each group
In this example, we are identifying the number of records by Crop type. There are two ways this can be accomplished:
dat %>%
filter(Information == "Yield (Hg/Ha)",
Year >= 2005 & Year <=2010,
Country=="United States of America") %>%
group_by(Crop) %>%
count()Or,
dat %>%
filter(Information == "Yield (Hg/Ha)",
Year >= 2005 & Year <=2010,
Country=="United States of America") %>%
group_by(Crop) %>%
summarise(Count = n())# A tibble: 7 x 2
Crop Count
* <chr> <int>
1 Barley 6
2 Buckwheat 6
3 Maize 6
4 Millet 6
5 Oats 6
6 Rye 6
7 Sorghum 6The former uses the count() function and the latter uses the summarise() and n() functions.
Summarize by mean yield and year range
Here’s another example where two variables are summarized in a single pipe.
dat.grp <- dat %>%
filter(Information == "Yield (Hg/Ha)",
Year >= 2005 & Year <=2010,
Country=="United States of America") %>%
group_by(Crop) %>%
summarise( Yield = mean(Value), `Number of Years` = max(Year) - min(Year))
dat.grp# A tibble: 7 x 3
Crop Yield `Number of Years`
* <chr> <dbl> <int>
1 Barley 35471. 5
2 Buckwheat 10418. 5
3 Maize 96151. 5
4 Millet 16548. 5
5 Oats 22619. 5
6 Rye 17132. 5
7 Sorghum 42258. 5Normalizing each value in a group by the group median
In this example, we are subtracting each value in a group by that group’s median. This can be useful in identifying which year yields are higher than or lower than the median yield value within each crop group. We will concern ourselves with US yields only and sort the output by crop type. We’ll save the output dataframe as dat2.
dat2 <- dat %>%
filter(Information == "Yield (Hg/Ha)",
Country == "United States of America") %>%
select(Crop, Year, Value) %>%
group_by(Crop) %>%
mutate(NormYield = Value - median(Value)) %>%
arrange(Crop)Let’s plot the normalized yields by year for Barley and add a 0 line representing the (normalized) central value.
plot( NormYield ~ Year, dat2[dat2$Crop == "Barley",] )
abline(h = 0, col="red")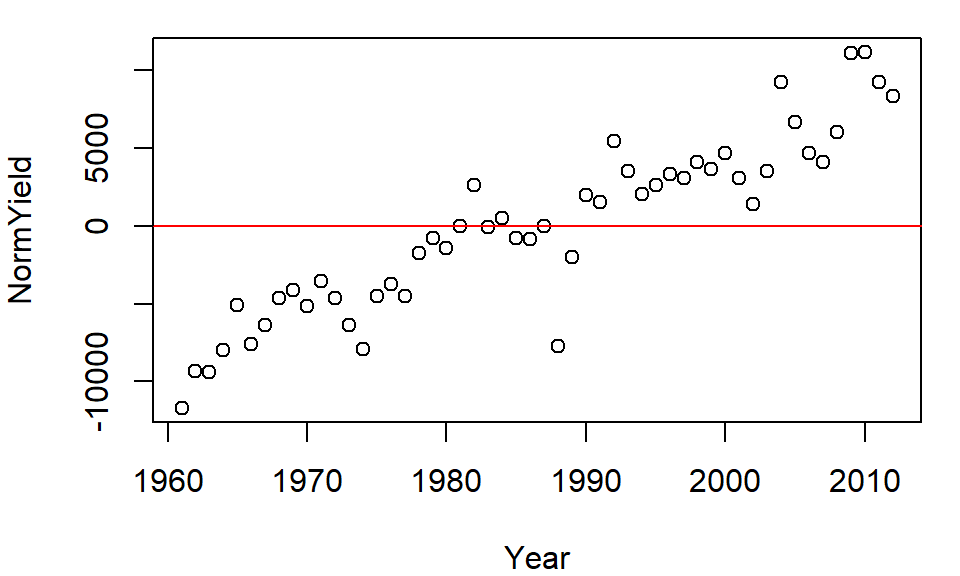
The relative distribution of points does not change, but the values do (they are re-scaled) allowing us to compare values based on some localized (group) context. This technique will prove very useful later on in the course when EDA topics are explored.
dplyr’s output data structure
Some of dplyr’s functions such as group_by/summarise generate a tibble data table. For example, the dat.grp object created in the last chunk of code is associated with a tb_df (a tibble).
class(dat.grp)[1] "tbl_df" "tbl" "data.frame"A tibble table will behave a little differently than a data frame table when printing to a screen or subsetting its elements. In most cases, a tibble rendering of the table will not pose a problem in a workflow, however, this format may prove problematic with some older functions. To remedy this, you can force the dat.grp object to a standalone dataframe as follows:
dat.df <- as.data.frame(dat.grp)
class(dat.df)[1] "data.frame"